
In biology, homology is commonly defined as any similarity between structures of organisms in different taxa that derives from similar structures in their shared ancestry. Examples of homology occur at different levels of organization. Entire anatomical structures that are similar in different biological taxa (species, genera, and so on) would be termed homologous if their common ancestor also exhibits a similar anatomical structure. Closely correlated partial sequences in DNA or protein could be similarly labeled if common ancestry is considered to be the cause.
Such a definition of homology is a redefinition from the classical understanding of the term, which predated Charles Darwin's theory of evolution, having been coined by Richard Owen in the 1840s. Historically, homology was defined as similarity in structure and position, such as the pattern of bones in a bat's wing and those in a porpoise's flipper (Wells, 2000). Historically, and still today, the term analogy has signified functional similarity, such as the wings of a bird and those of a butterfly.
Darwin claimed homology in the classical sense as evidence of evolution by common descent (theory of descent with modification) under the assumption that new forms develop on the foundation of earlier stages. His evidence included the similar skeletal structures (utilizing same bones) of the forelimbs of humans, bats, birds, cats, and porpoises.
The discovery, however, of clear exceptions to Darwin's premise meant that structural similarity could no longer be used as evidence of a common ancestor. One such exception is the remarkable structural similarity between the eyes of humans and octopuses, two organisms whose remote common ancestor is not thought to have had such eyes (Ogura, Ikeo, and Gojobori, 2004). If shared ancestry must be established apart from structural similarity before structures can be called homologous, then using two structures' similarities as evidence of their evolutionary origin, including their common ancestry, would entail the use of circular reasoning.
The word homologous derives from the ancient Greek ομολογειν, “to agree.” The term homology is also used in a non-evolutionary sense in terms of homologous chromosomes, meaning a pair of non-identical chromosomes from a diploid organism that can pair (synapse) during meiosis, or regions of chromosomes with the same set of genes.
Homology of structures in evolution
Homology in the classical sense, as similarity in structure and position of anatomical features between different organisms, was important evidence used by Darwin. However, it has now been redefined as structures that trace to common ancestors. Mayr (1982) states:
After 1859 there has been only one definition of homologous that makes biological sense… Attributes of two organisms are homologous when they are derived from an equivalent characteristics of the common ancestor.
Homology is different from analogy. For example, the wings of insects, the wings of bats, and the wings of birds are analogous but not homologous; this phenomenon is called "homoplasy." These similar structures evolved through different developmental pathways, in a process labeled as convergent evolution.
Wells (2000) notes that homology as now defined can no longer be used as evidence for evolution because of circular reasoning. Furthermore, he finds even retaining the pre-Darwinian definition of homology as structural similarity problematic because of such factors as the lack of correspondence between homology and development pathways, since there are examples of similar structures in adult forms where ancestry is assumed that actually are products of radically different development processes. Likewise, he notes that some assumed homologous structures trace to different genes.
Homology: Pentadactyl limb
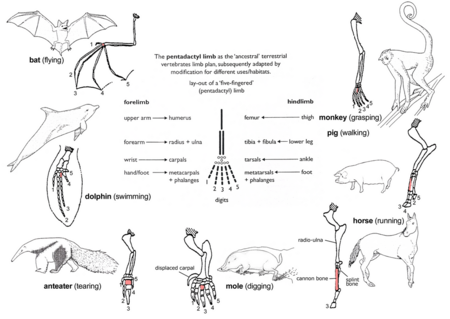
The pattern of limb bones known as a pentadactyl limb is an example of homologous structures. This pattern is found in all classes of tetrapods (i.e. from amphibians to mammals). It can even be traced back to the fins of certain fossil fish from which the first amphibians are thought to have evolved. The limb has a single proximal bone (humerus), two distal bones (radius and ulna), a series of carpals (wrist bones), followed by five series of metacarpals (palm bones) and phalanges (digits). Throughout the tetrapods, the fundamental structures of pentadactyl limbs are the same, indicating that they originated from a common ancestor. But it is considered that in the course of evolution, these fundamental structures have been modified. They have become superficially different and unrelated structures in order to serve different functions in adaptation to different environments and modes of life. This phenomenon is clearly shown in the forelimbs of mammals. For example:
- In the monkey, the forelimbs are much elongated to form a grasping hand for climbing and swinging among trees.
- In the pig, the first digit is lost, and the second and fifth digits are reduced. The remaining two digits are longer and stouter than the rest and bear a hoof for supporting the body.
- In the horse, the forelimbs are adapted for support and running by great elongation of the third digit bearing a hoof.
- The mole has a pair of short, spade-like forelimbs for burrowing.
- The anteater uses its enlarged third digit for tearing down ant hills and termite nests.
- In the whale, the forelimbs become flippers for steering and maintaining equilibrium during swimming.
- In the bat, the forelimbs have turned into wings for flying by great elongation of four digits, and the hook-like first digit remains free for hanging from trees.
Homology: Insect mouthparts

In insects,the basic structures of the mouthparts are the same, including a labrum (upper lip), a pair of mandibles, a hypopharynx (floor of mouth), a pair of maxillae, and a labium. These structures are enlarged and modified; others are reduced and lost. The modifications enable the insects to exploit a variety of food materials:
(A) Primitive state—biting and chewing: E.g., grasshopper. Strong mandibles and maxillae for manipulating food.
(B) Ticking and biting: E.g., honeybee. Labium long to lap up nectar; mandibles chew pollen and mold wax.
(C) Sucking: E.g., butterfly. Labrum reduced; mandibles lost; maxillae long forming sucking tube.
(D) Piercing and sucking: E.g, female mosquito. Labrum and maxillae form tube; mandibles form piercing stylets; labrum grooved to hold other parts.
Homology: Other arthropod appendages
Insect mouthparts and antennae are considered homologues of insect legs. Parallel developments are seen in some arachnids: The anterior pair of legs may be modified as analogues of antennae, particularly in whip scorpions, which walk on six legs. These developments provide support for the theory that complex modifications often arise by duplication of components, with the duplicates modified in different directions.
Homology of sequences in genetics
In genetics, homology is measured by comparing protein or DNA sequences, and homologous genes share a high sequence identity or similarity, supporting the hypothesis that they share a common ancestor. Sequence homology may also indicate common function. Homologous chromosomes are non-identical chromosomes that can pair (synapse) during meiosis, and are believed to share common ancestry.
Homology among proteins and DNA is often concluded on the basis of sequence similarity, especially in bioinformatics. For example, in general, if two genes have an almost identical DNA sequence, it is likely that they are homologous. But sequence similarity may arise from different ancestors: Short sequences may be similar by chance, and sequences may be similar because both were selected to bind to a particular protein, such as a transcription factor. Such sequences are similar but not homologous. Sequence regions that are homologous are also called conserved.
The phrase "percent homology," is sometimes used but is incorrect. "Percent identity" or "percent similarity" should be used to quantify the similarity between the biomolecule sequences. For two naturally occurring sequences, percent identity is a factual measurement, whereas homology is a hypothesis supported by evidence. One can, however, refer to partial homology where a fraction of the sequences compared (are presumed to) share descent, while the rest does not.
Many algorithms exist to cluster protein sequences into sequence families, which are sets of mutually homologous sequences.
There are two types of homology of sequences: Orthologous and paralogous.
Orthology
Orthologs, or orthologous genes, are any genes in different species that are similar to each other and are attributed to have originated from a common ancestor, regardless of their functions. Thus, orthologs are separated by an evolutionary speciation event: If a gene exists in a species, and that species diverges into two species, then the divergent copies of this gene in the resulting species are orthologous. The term "ortholog" was coined in 1970.
A second definition of orthologous has arisen to describe any genes with very similar functions in different species. This differs from the original definition in that there is no statement about evolutionary relation, or similarity in sequence or structure.
Orthologous sequences provide useful information in taxonomic classification studies of organisms. The pattern of genetic divergence can be used to trace the relatedness of organisms. Two organisms that are very closely related are likely to display very similar DNA sequences between two orthologs. Conversely, an organism that is further removed evolutionarily from another organism is likely to display a greater divergence in the sequence of the orthologs being studied.
Paralogy
Homologous sequences are paralogous if they were separated by a gene duplication event; if a gene in an organism is duplicated to occupy two different positions in the same genome, then the two copies are paralogous.
A set of sequences that are paralogous are called paralogs of each other. Paralogs typically have the same or similar function, but sometimes do not. It is considered that due to lack of the original selective pressure upon one copy of the duplicated gene, this copy is free to mutate and acquire new functions.
Paralogous sequences provide useful insight to the way genomes evolve. The genes encoding myoglobin and hemoglobin are considered to be ancient paralogs. Similarly, the four known classes of hemoglobins (hemoglobin A, hemoglobin A2, hemoglobin S, and hemoglobin F) are considered to be paralogs of each other. While each of these genes serves the same basic function of oxygen transport, they differ slightly in function: fetal hemoglobin (hemoglobin F) has a higher affinity to oxygen than adult hemoglobin.
Another example can be found in rodents such as rats and mice. Rodents have a pair of insulin genes considered to be paralogous, although it is unclear if any divergence in function has occurred.
Paralogous genes often belong to the same species, but this is not necessary. For example, the hemoglobin gene of humans and the myoglobin gene of chimpanzees are considered paralogs. This is a common problem in bioinformatics; when genomes of different species have been sequenced and homologous genes have been found, one can not immediately conclude that these genes have the same or similar function, as they could be paralogs whose function has diverged.
Xenology
Homologs resulting from horizontal gene transfer between two organisms are termed xenologs. Xenologs can have different functions, if the new environment is vastly different for the horizontally moving gene. In general, though, xenologs typically have similar function in both organisms (NCBI, 2004).
Homologous chromosome sets
Homologous chromosomes are defined as non-identical chromosomes that can pair (synapse) during meiosis (King and Stansfield, 1997). Except for the sex chromosomes, homologous chromosomes share significant sequence similarity across their entire length, typically contain the same sequence of genes, and pair up to allow for proper disjunction during meiosis. The chromosomes can also undergo cross-over at this stage. There may be some variations between genes on homologs giving rise to alternate forms or alleles. Sex chromosomes have a shorter region of sequence similarity. Based on the sequence similarity and human knowledge of biology, it is believed that they are paralogous.
ReferencesISBN links support NWE through referral fees
- DePinna, M. C. C. 1991. Concepts and tests of homology in the cladistic paradigm. Cladistics 7: 367-394.
- Dewey, C., and L. Pachter. 2006. Evolution at the nucleotide level: The problem of multiple whole-genome alignment. Human Molecular Genetics 15(Review Issue 1): R51-R56. Retrieved April 26, 2007.
- Fitch, W. M. 2000. Homology: A personal view on some of the problems. Trends Genet. 16(5): 227-31. Retrieved April 26, 2007.
- Gegenbaur, G. 1898. Vergleichende Anatomie der Wirbelthiere. Leipzig.
- Haeckel, Е. 1866. Generelle Morphologie der Organismen. Bd 1-2. Вerlin.
- King, R. C., and W. D. Stansfield. 1997. A Dictionary of Genetics, 5th ed. New York: Oxford University Press. ISBN 0195094417.
- National Center for Biotechnology Information (NCBI). 2004. Just the facts: A basic introduction to the science underlying NCBI resources: Systematics and molecular phylogenetics. NCBI: A Science Primer.
- Mayr, E. 1982. The Growth of Biological Thought. Cambridge, MA: Harvard University Press. ISBN 0674364457.
- Ogura, A., Ikeo, K., Gojobori, T. 2004. Comparative Analysis of Gene Expression for Convergent Evolution of Camera Eye Between Octopus and Human. Genome Res. 14: 1555-1561. Retrieved January 28, 2009.
- Owen, R. 1847. On the Archetype and Homologies of the Vertebrate Skeleton. London.
- Wells, J. 2000. Icons of Evolution. Washington, D.C.: Regnery Publishing. ISBN 0895262762.
Credits
New World Encyclopedia writers and editors rewrote and completed the Wikipedia article in accordance with New World Encyclopedia standards. This article abides by terms of the Creative Commons CC-by-sa 3.0 License (CC-by-sa), which may be used and disseminated with proper attribution. Credit is due under the terms of this license that can reference both the New World Encyclopedia contributors and the selfless volunteer contributors of the Wikimedia Foundation. To cite this article click here for a list of acceptable citing formats.The history of earlier contributions by wikipedians is accessible to researchers here:
The history of this article since it was imported to New World Encyclopedia:
Note: Some restrictions may apply to use of individual images which are separately licensed.